Seurat CCA? It's just a simple extension of PCA!
Introduction
The canonical correlation analysis (CCA) implemented as part of Seurat software package is one of the most popular methods for batch effects correction in single-cell RNA-seq datasets. However, the Method part of the original paper Comprehensive Integration of Single-Cell Data is not easy to read. After carefully reading the math, and also doing some experiments, my friend (Ziyu Chen) and I realized that the math behind the so-called CCA method actually is better described by the dual form of PCA (“dual PCA” for short). This understanding provides more clarity into the objective function solved by the “Seurat CCA algorithm” and also, as I describe later, provides some insights to improve it.
In this blog,
-
I will first introduce PCA and dual PCA. While PCA/dual PCA computes the self-similarity in one dataset (\(XX^T\)), we can extend them to compute the similarity across two datasets (\(XY^T\)), and this is equivalent to “Seurat CCA”!
-
Importantly, this derivation clarifies that “Seurat CCA” is missing the multiplication with singular values in the embedding process that dual PCA implies. We demonstrated by experiments that if the singular values are considered (as they should be), more biological variation will be preserved.
-
Finally, it’s not difficult to show that there is an intrinsic connection between the two most popular batch-effect removing methods, namely “Seurat CCA” and MNN (Mutual nearest neighbor).
Principal Components Analysis (PCA)
PCA is a very popular method for dimension reduction.
Direct PCA
PCA aims to project data points \(x \in R^{g}\) into a linear subspace \(R^k\) while preserving as much as variability as possible.
For a given dataset \(X\in R^{n \times g}\) (In single cell RNAseq data, \(n\) is the number of cells, \(g\) is the number of genes), PCA finds a linear embedding \(Z= XV \in R^{k \times n}\), where \(X\in R^{n\times g}, V\in R^{g \times k}\), to represent \(X\) while preserving as much as possible the variability in \(X\). (Note: An equivalent and another common definition of PCA is the projection onto a subspace minimizing the squared reconstruction error.)
The solution \(V\) can be calculated from the singular value decomposition (SVD) of \(X\):
\[X = U\Sigma V^T, X^TX=V \Sigma^2 V^{T}.\]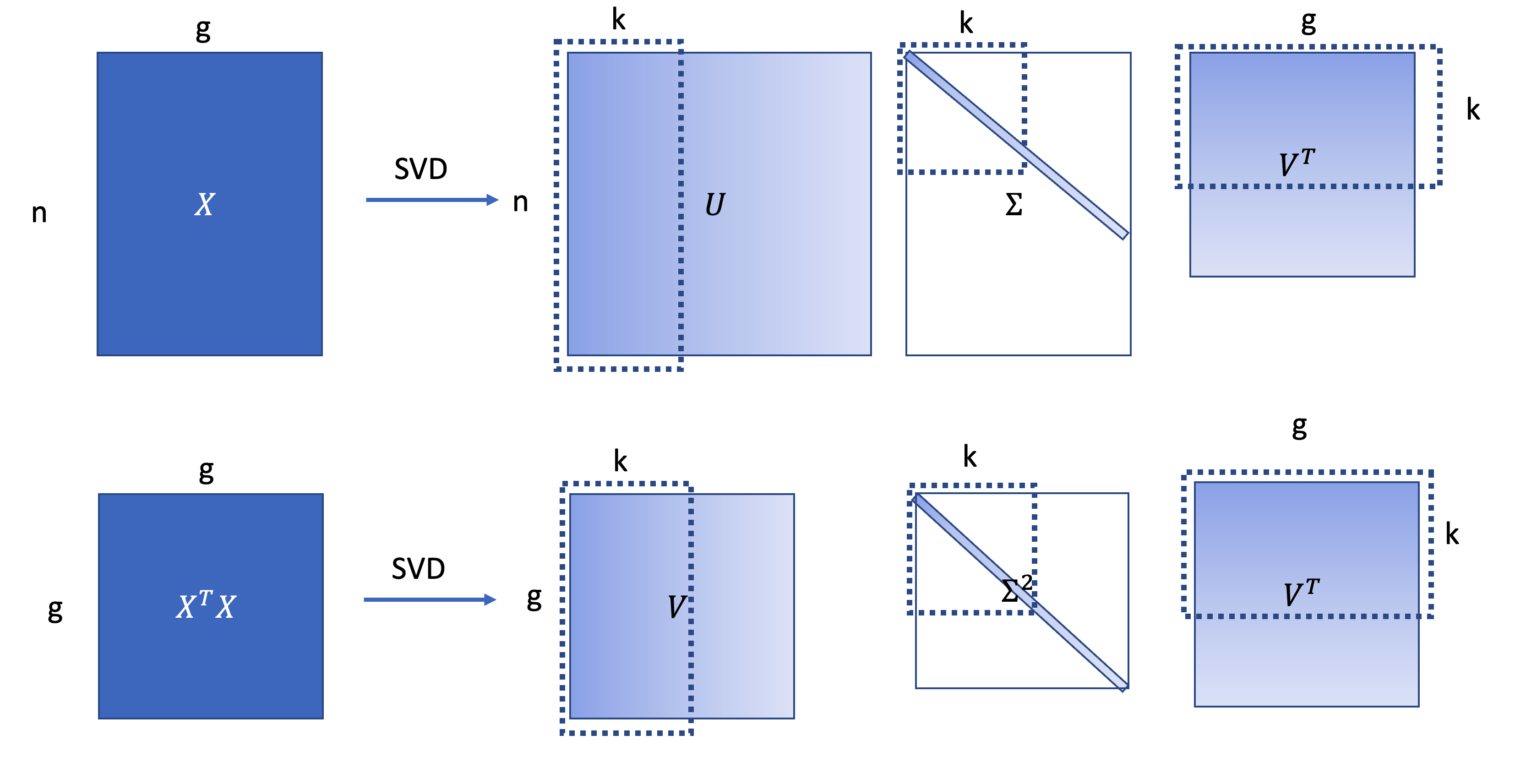
\(X\in R^{n\times g}\) is the gene-gene expression matrix. \(X^TX \in R^{g\times g}\) are the gene co-variance matrix. Equivalently put, the SVD provides the best low rank approximation of gene-gene co-variance matrix as the multiplication of top \(K\) singular values and vectors of \(X\) shown below:
And the embedding we are looking for in PCA will be
\[Z = XV_{1:g, 1:k} =U\Sigma V^T V_{1:g,1:k} = U_{1:n, 1:k}\Sigma_{1:k}.\]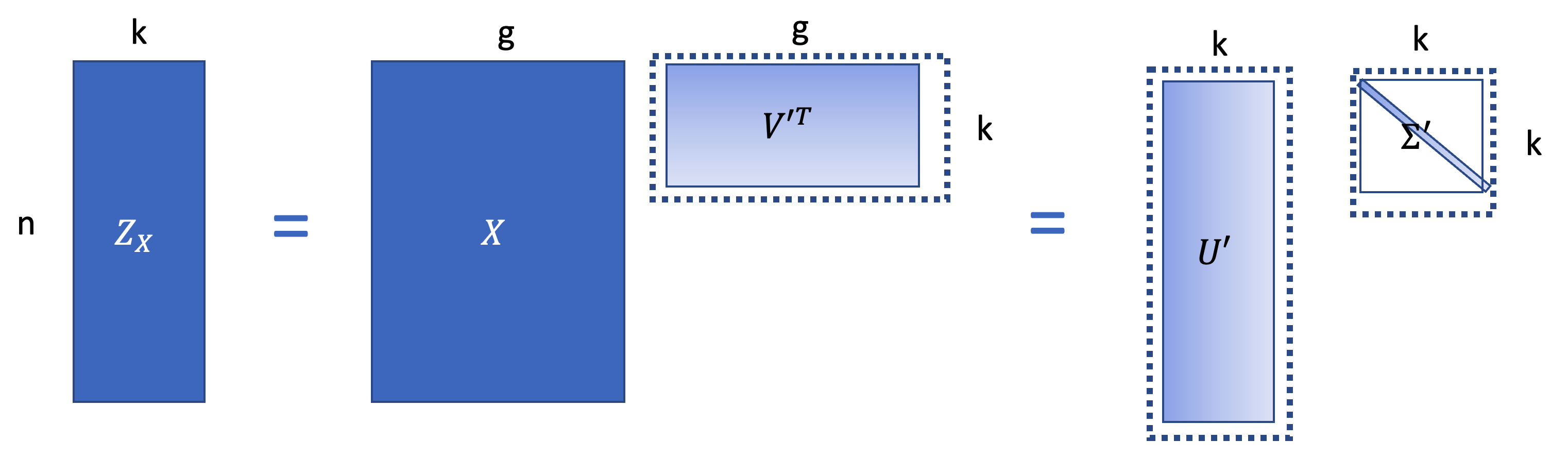
\(Z_X \in R^{n\times k}\) is the low dimensional embeddings. } \(The V^{'T} \in R^{g\times k}\) is the project matrix.Then based on the SVD, \(Z = U_{1:n, 1:k}\Sigma_{1:k}\)
Dual PCA
In the above, SVD was applied to \(X^TX\), which gives us a low rank approximation to the full gene-gene covariance matrix. But what if we apply SVD to \(XX^T\)? This would be applying SVD in the “dual space”, which would give us the best low rank approximation to sample-sample covariance matrix
\[X = U\Sigma V^T, XX^T=U\Sigma^2 U^{T}\]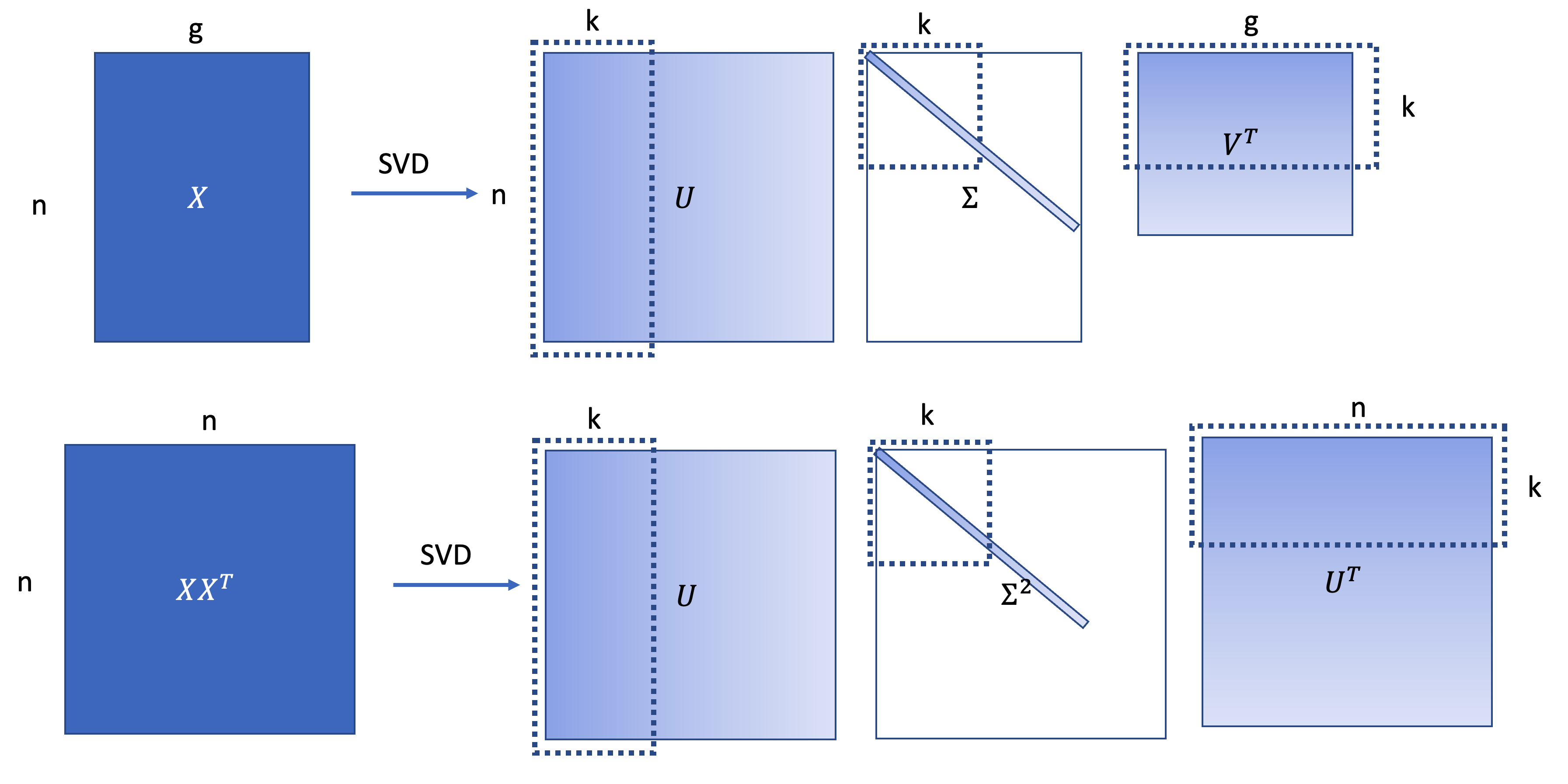
When applying this “dual PCA” to the gene expression matrix as formulated above, The SVD provides the best low rank approximation of cell-cell covariance matrix by select the Top \(k\) singular value and the vectors.
And \(Z = U_{1:n, 1:k}\Sigma_{1:k}\) will be the embedding, which is the same as what we derived in PCA.
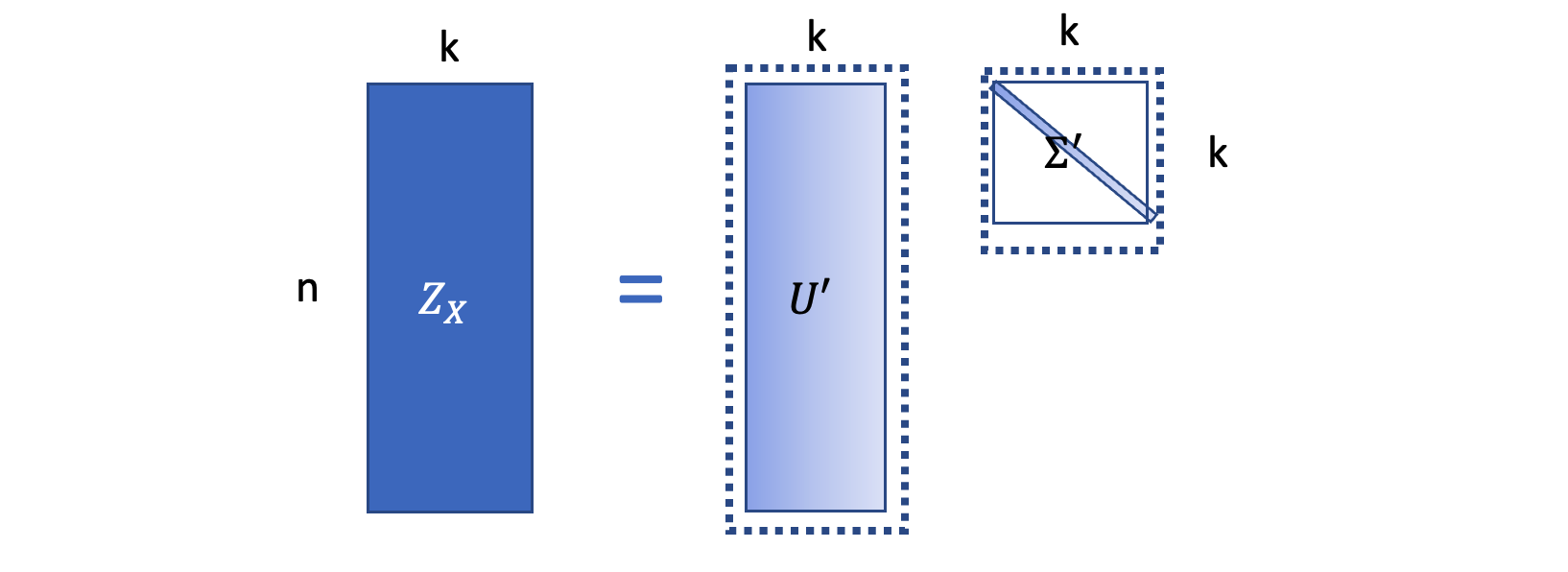
\(Z_X \in R^{n\times k}\) is the low dimensional embeddings. Then based on the SVD, \(Z = U_{1:n, 1:k}\Sigma_{1:k}\) is the best embedding which approximate the similarity matrix best.
We can check it by
\[|XX^T - ZZ^T| = |U\Sigma^2 U^{T} - U_{1:n, 1:k}\Sigma_{1:k}^2 (U_{1:n,1:k})^T|.\]Here, the eigenvalues in the diagonal matrix \(\Sigma\) is monotonically decreasing, meaning that \(U_{1:n, 1:k}\Sigma_{1:k}^2 (U_{1:n,1:k})^T\) is the best low-rank approximation of \(U\Sigma^2 U^{T}\).
“Seurat CCA” – an Extension of Dual PCA
In this section, we will derive an extension of dual PCA and show that it’s almost the same as the “Seurat CCA” method.
The dual PCA is formulated to find a low-dimensional embedding while preserving the similarity between samples in one dataset. But how do we apply it to preserve similarities when we have two datasets?
More formally, given two datasets(e.g. from two batches), \(X\in R^{n\times g}\) and \(Y\in R^{m\times g}\), where \(n\) and \(m\) means the number of cells, \(g\) means the number of genes. The goal is to find two low-dimensional linear embeddings for both datasets \(Z_X\in R^{n\times k},Z_Y\in R^{m \times k}\) to preserve the similarity matrix between cells from different datasets rather than in a single dataset. Mathematically, we can formulate this as follows: we’d like to find an embedding that minimizes the difference between “true” similarity matrix and the similarity of their embedding, \(\|Z_XZ_Y^T - XY^T\|\), which also means finding a low rank approximation of \(XY^T\).
Specifying the goal in this way, it may now be more clear that a small modification of dual PCA can solve the for the desired embeddings – In dual PCA, we apply SVD to \(XX^T\), but now we can apply the SVD to \(XY^T\). As in Dual PCA, the embedding preserves the sample similarity within one dataset, in this new case, the embeddings will preserve the cross-batch sample similarity.
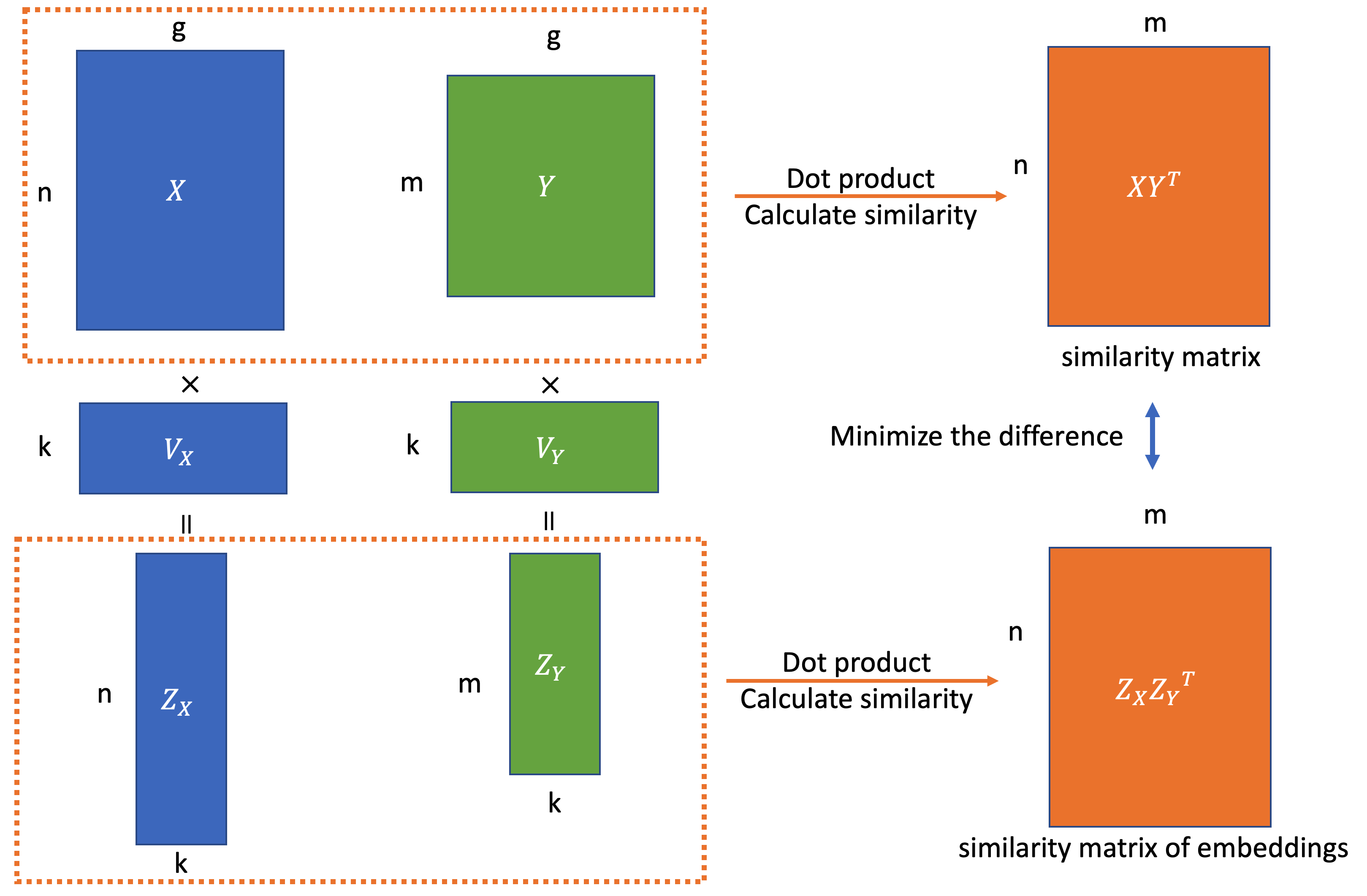
\(X\in R^{n\times g}, Y\in R^{m\times g}\) are the gene expression matrix. \(XX^T \in R^{n\times n}, YY^T \in R^{m\times m}\) are the cell-cell similarity matrixes. \(V_X, V_Y\in R^{g\times k}\) are the project matrix for these two batches. \(Z_X \in R^{n\times k}, Z_Y \in R^{m\times k}\) are the low dimensional embeddings(projection) of two batch data \(X, Y\). The objective function can be written to find these embeddings which preserve the cell-cell similarity matrix as much as possible.
We can get
\[XY^T = U\Sigma V^T\]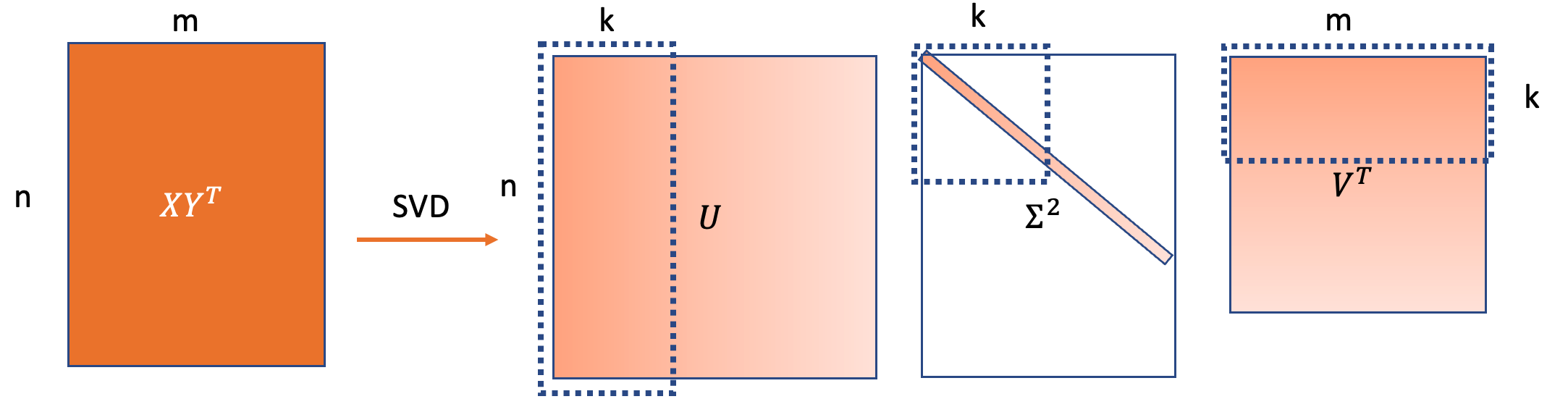
The best embeddings will be
\[Z_X = U_{1:n,1:k}(\Sigma_{1:k})^\frac{1}{2}, Z_Y = V_{1:n, 1:k}(\Sigma_{1:k})^\frac{1}{2}\]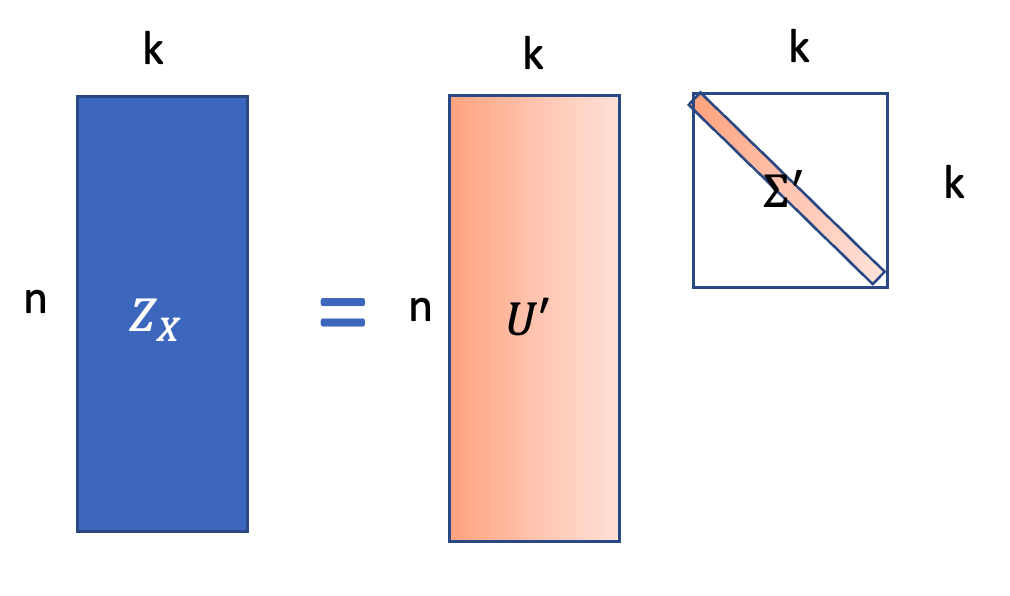
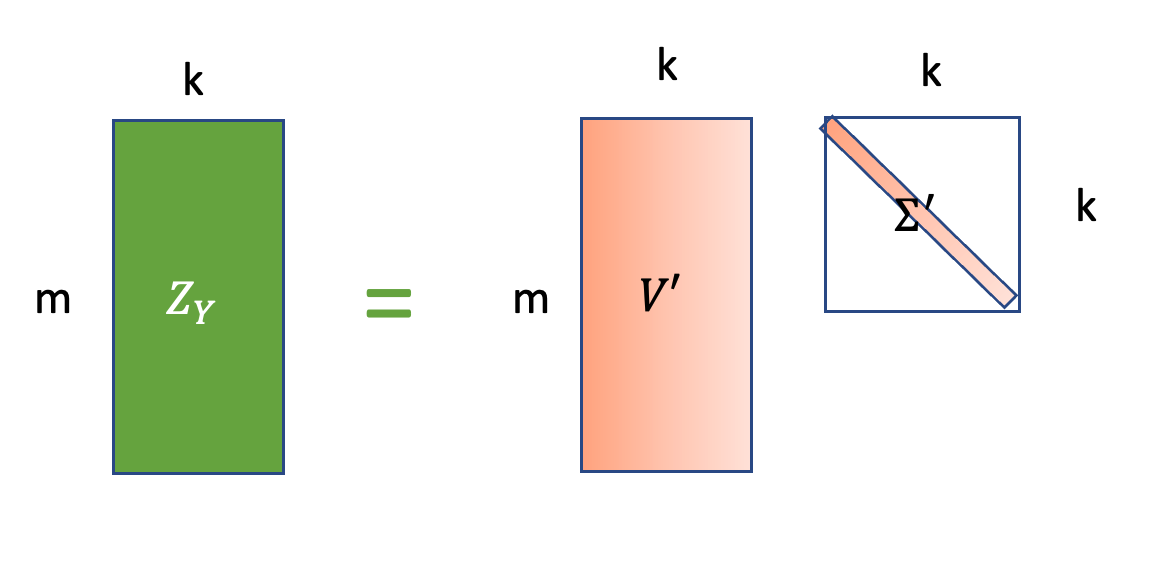
\(Z_X \in R^{n\times k}, Z_Y \in R^{m\times k}\) are the best low dimensional embeddings(projection) of two batch data \(X, Y\).
We can also check this approximation:
\[\begin{aligned} |XY^T - Z_XZ_Y^T| &= | U\Sigma V^T - U_{1:n,1:k}\Sigma_{1:k}^\frac{1}{2} (V_{1:n, 1:k}\Sigma_{1:k}^\frac{1}{2})^T| \\ &= | U\Sigma V^T - U_{1:n,1:k}\Sigma_{1:k} (V_{1:m, 1:k})^T| \end{aligned}\]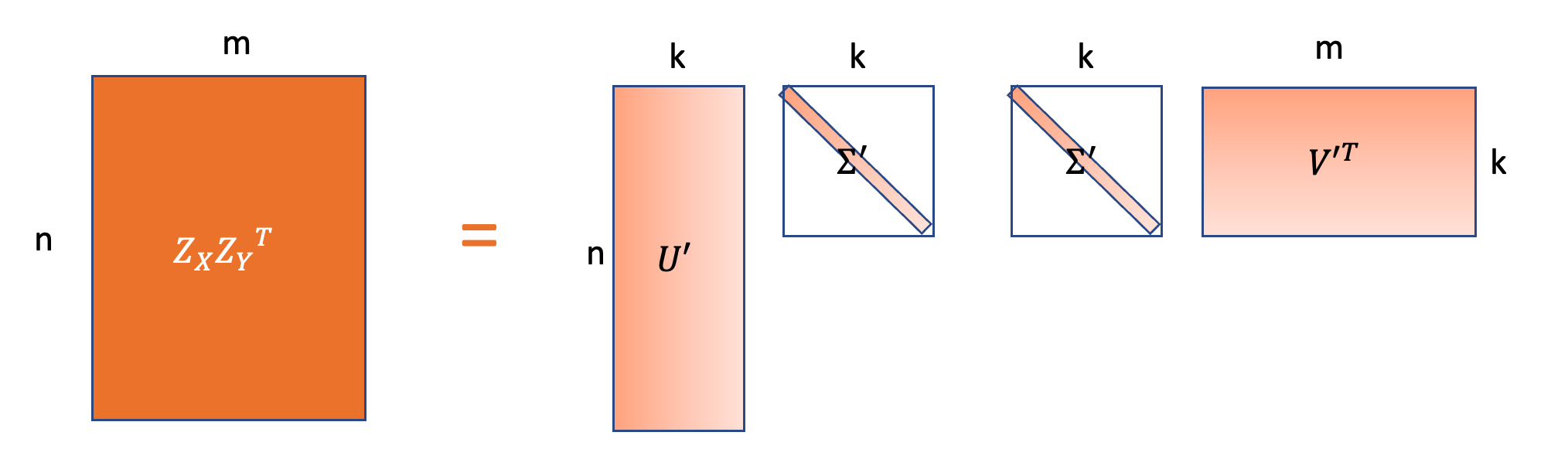
\(Z_XZ_Y^T\) is the similarity matrix based on the low dimensional embeddings.
Based on SVD, \(U_{1:n,1:k}\Sigma_{1:k} (V_{1:m, 1:k})^T\) is the best low-rank approximation of \(U\Sigma V^T\).
Summary of Math
Dual PCA
- Data: \(X\in R^{n\times g}\), \(n\) is the number of cells, \(g\) is the number of genes.
- Task: \(Z\) to represent the data \(X\).
- Object: Minimize \(\|ZZ^T - XX^T\|\) (Dual PCA).
- Dual PCA Solution: \(XX^T= U\Sigma^2U^T\), \(Z = U_{1:n,1:g}\Sigma_{1:k}\).
Dual PCA Extended to Two Datasets
- Data: \(X\in R^{n\times g}, Y\in R^{m \times g}\), \(n\) and \(m\) are the number of cells, \(g\) is the number of genes
- Task: \(Z_X\in R^{n\times k}\) and \(Z_Y\in R^{m \times k}\) to represent the two datasets \(X\) ane \(Y\).
- Object: Minimize \(\|Z_XZ_Y^T - XY^T\|\) .
- Solution: \(XY^T = U\Sigma V^T\), \(Z_X = U\Sigma^\frac{1}{2}, Z_Y = V \Sigma^\frac{1}{2}\)
Back to the “Seurat CCA” paper
In the Method section of the “Seurat CCA” paper, authors had several assumptions to get to the final result of cell embeddings. An important assumption is to “treat the covariance matrix within each dataset as diagonal”, meaning that genes are independent to each other, which is NOT true.

Alternatively, applying SVD to \(XY^T\) is intuitive and natural - it is just to capture the cell similarity across batches and does not need any assumption.
Furthermore, in the original paper, the cell embeddings of two datasets are \(Z_X = U\) and \(Z_Y = V\), is missing the multiplication with the singular value values, as would be implied by SVD application to \(XY^T\)
From method part of the original paper. In the original paper, \(X\in R^{g\times n}, Y\in R^{g\times m}\), just a different notation.
To compare the difference in algorithm performance with or without the singular value, we implemented these two methods to data from Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nature biotechnology (2020)
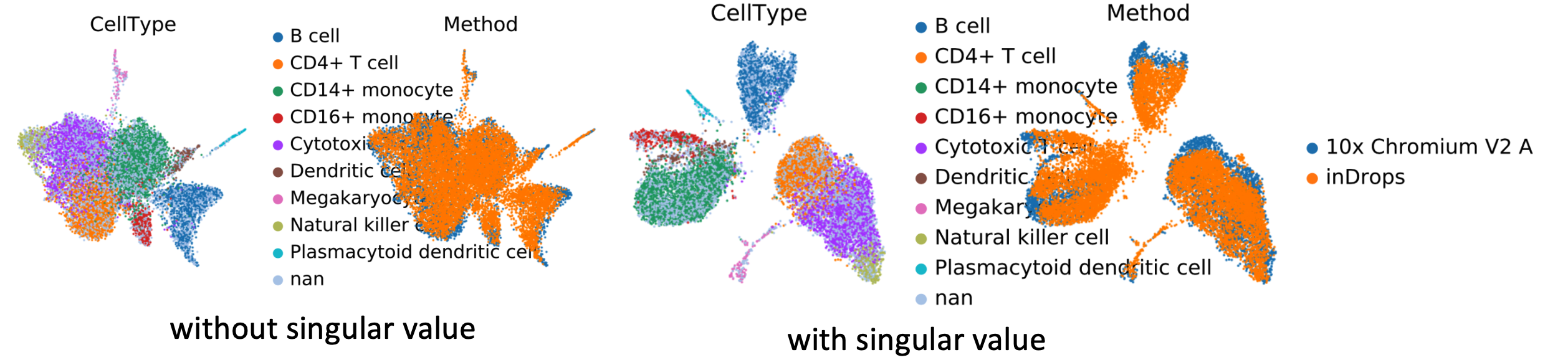
As shown in the plot above, when we multiply the embedding by singular values the different cell types are more separated from each other in Umap visualization, meaning that more biological variation is preserved in the embedding.
Difference between “Seurat CCA” and Real CCA
The “Seurat CCA” is taking the projection vector from the traditional CCA directly as cell embeddings. But in fact, the classical definition of CCA would imply projecting genes into a common space rather than cells.
Connection with the Mutual Nearest Neighbor (MNN) method.
There is an intrinsic connection between “Seurat CCA” and MNN, another popular method in removing batch-effect.
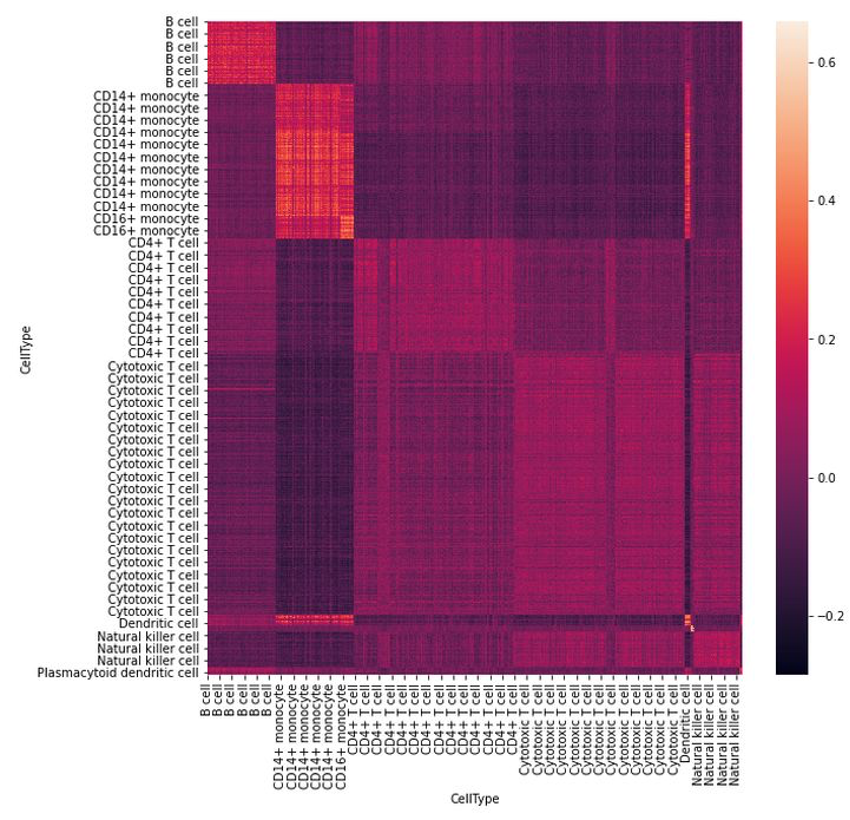
“Seurat CCA” has the assumption that biologically more similar cells from different batches have a higher mathematical similarity (i.e. the dot product), and similarly, MNN assume similar cells from different batches have smaller Euclidean distance defined in the algorithm. In fact, if one standardizes the data (so that each cell has mean of 0 and STD of 1), dot product and euclidean distance are equivalent.
The above assumptions are clearly observable in “real data”. We can plot a heatmap of the similarity matrix of two different batches, and what we see is a higher similarity along the diagonal.
Conclusion
-
Based on our understanding, the math behind the “Seurat CCA” algorithm is technically closer to a dual PCA formulation.
-
In the original paper, the assumption that the covariance matrix of gene expression is diagonal, is not necessary.
-
Furthermore, considering the formulation to preserve the most similarity (dual PCA), the low-dimensional cell embeddings should multiply the singular value, which is currently missing in the “Seurat CCA” algorithm.
-
And finally, there is an intrinsic connection between MNN and “Seurat CCA” (extended dual PCA).
Acknowledge
Thanks for my advisor Sara’s revision and my girl friend Jiahui Peng’s help on the blog writing. Thanks for Ziyu Chen, Zhijie Cao, Weixu Wang’s discussion and comments.
If you like this blog, you can give me a like on twitter! Thanks!
Hi all, I want to share my first blog - "Seurat CCA? It's just a simple extension of PCA!"https://t.co/K2mUTOkeGT#Bioinformatics #singlecell
— Xinming Tu (@TuXinming) April 11, 2022